|
Pathway Plot Manual © Anders Broe Bendtsen 15 March 1999 What does the program
do?
Pathway
Plot is a program, which automatically will plot the dominant reaction paths,
in a detailed chemical kinetic model, based on simulation output from a
SENKIN simulation. Pathway Plot is a
postprocessor to SENKIN. From one or more initial key species and a binary
output file from a SENKIN simulation, the program defines the set of
important key species and plots the significant reactions between these
species. You may choose to view only species containing a defined element,
and at any time you may take over control of the plots. Installation
Click
here to register for Downloading PP_INST.EXE The downloaded file will be 800K.
Run PP_INST.EXE This will prompt you for
a new directory and unpack PP.EXE, an example CKLINK and SAVE.BIN and a required SALFLIBC.DLL system file. For information the ASCII
mechanism, CK.MEC and SENKIN input and output SK.INP and SK.OUT are also unpacked to the
directory, but these files are not needed. |
|
Pathway Plot manual
Running Pathway Plot
First run your SENKIN simulation to generate a SAVE.BIN binary output
file.
Ensure that the binary
SAVE.BIN and CKLINK. files of the reaction set is in the same directory as
PP.EXE
If your binary files have
other names use the DOS command SET as follows:
SET CHEMKIN=CHEM.BIN
(for defining the binary file of the reaction set to be "CHEM.BIN")
SET SENKIN=SENKIN.BIN
(for defining the binary file of the SENKIN simulation to be
"SENKIN.BIN")
Call PP.EXE
If you do not have a
default setup (found in a ROPCONT.INP file) you will be prompted to setup Pathway
Plot. You may select Setup from the File menu to obtain the same dialogue
boxes. You must select one key species, and subsequently press Continue.
When the set-up is
sufficiently defined, you must press the OK button to see the Pathway Plot of
the next time step. Unmark Pause in the File menu to see a continuous
"movie" of the reaction.
To exit select Exit from
the File menu.
Pathway Plot's display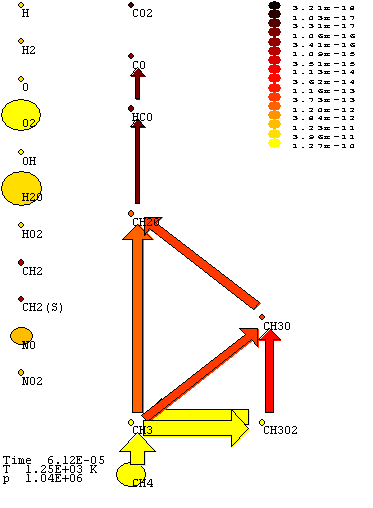
In the
centre of the display, will be the Pathway Plot, with arrows between species. The
width and colour of arrows will be proportional to the activity of the path
between the species. The species will be followed by a circle. The colour of
the circle will correspond to the total reaction of the species, and the size
of the circle to the logarithm of the concentration of the species.
To the right on the plot is
a scale translating the colours of arrows and circles to rates of progress
(units are defined in your CHEMKIN-II reaction set). The rate of progress is
the net production (or removal) of a species through a reaction, but for a
Pathway Plot arrow from species A to P
Introduction it may be the sum of several
reaction, with one reactant (A) and product (P) in common.
To the left are the active
species which have been defined as unwanted on the plot.
In the top left corner is
an OK button which is used to show the pathway plot at the next time step.
Pathway Plot setup
In setup
you choose which species to display and you define the display here. Setup is
chosen from the File menu. The setup window has three parts, "Key
Species", "Drawing" and "Position of species" as
defined below.
To continue plotting from the setup, select 'Continue'. If no species are
selected you will prompted if you want to exit.
Selection of key species
You will be
prompted for an initial set of key species. Select by clicking the species.
To avoid cluttering of your
display you may choose to select some "Ignored" species. If you do
not want Pathway Plot to expand your set of active species, remove the mark in
the box next to "Expand key species".
If you want to start from
scratch you may want to clear the selection of key species and/or ignored species.
To see species which must
contain a single element select this element in the active element box. This
will force all species not containing the active element to be ignored. You may
select several active species, and you may subsequently deselect the
"active elements" and manually select or deselect igored species.
Drawing of pathways
This setup
makes it possible to select the active range of your pathway plots. You may
define the colour range of your plot, and you may select the Relative production/removal
limit, which is the level of contribution needed for an arrow to be included
(and thus for expansion of species).
You may also choose how the reactions should be described; without text, by
reactants and products or by reaction numbers. You may select a colour scale
(Note that the flame scale is best for black and white printers).
Position of species
Pathway
Plot attempts to position the species, but at better plot may be obtained by
manually moving the species.
This may be done in the
setup, but a simpler method will be by clicking on a species. This will bring
up two open circles around the species, and a number of closed circles in the
available positions. Click on a closed circle to move the species. If you
regret moving the species click on the original position of the species.
The benefit of moving species in the setup is the possibility to exactly define
the position.
Working with saved setups
You may
save the setup by selecting Save or Save Default in the setup box. Save Default
will force the name to be ROPCONT.INP while Save will allow you select any
name. If a file is called ROPCONT.INP in the directory PP.EXE will use this
setup.
Load will allow you to
select a saved setup. Format of setup files
Printing
To print
the Pathway Plots, you must select Print from the File menu. This will have the
inadvertent effect of clearing the clipboard.
Alternatively you may
choose to copy the plot to a drawing program and modify the plot there. You do
this by selecting "Copy" from the menu. The Pathway Plots are copied
as vector drawings, allowing you full freedom in modification of the plots.
Tips for trouble shooting
File
name errors
Your output files must be named CKLINK. and SAVE.BIN
If your files have other names use the DOS command SET as follows:
SET CHEMKIN=CHEM.BIN
(for defining the binary file of the reaction set to be "CHEM.BIN")
SET SENKIN=SENKIN.BIN
(for defining the binary file of the SENKIN simulation to be
"SENKIN.BIN")
No expansion of species
set is seen
Select Expand key Species in Setup
If you have only one species select manual limit definitions
Is the Relative Limit set too low?
Program Requirements
Windows 95
or Windows NT 4.0 (not tested with Windows 98).
Binary output from SENKIN,
compiled with CKLIB vers. 3.3.
CKLINK. and SAVE.BIN file
from SENKIN-simulation (output from sensitivity analysis not needed)
If your files have other
names use the DOS command SET as follows:
SET
CHEMKIN=CHEM.BIN
(for
defining the binary file of the reaction set to be "CHEM.BIN")
SET SENKIN=SENKIN.BIN
(for
defining the binary file of the SENKIN simulation to be "SENKIN.BIN")
Preferably 65536 colour
high resolution display, but 640 by 480 in 256 colours is acceptable.
Format of ROPCONT.INP
|
Keyword |
Explanation |
|
KSPEC CH4
CO2 |
! Defines
key species to be plotted |
|
ISPEC OH
O H |
! Ignores
specific species |
|
ELEM
C |
! Ignores
all species without C |
|
POSI CH4
0.5 0.0 |
!
Positions CH4 in the centre at the bottom (x=0.5, y=0) |
|
POSI CO2
0.5 1.0 |
!
Positions CO2 in the centre at the top (x=0.5, y=1) |
|
LEVL CH4
1 |
! Put CH4
at the bottom |
|
LEVL CO2
10 |
! Put CO2
at the top |
|
MINF
1E-12 |
! Scaling
of flux'es. Minimum
1E-12 |
|
MAXF 1E-8 |
! Scaling of flux'es. Maximum 1E-12 |
POSI and LEVL keywords may
be used for all species in the mechanism, also ignored species. If species not
included in the mechanism are listed an error will be seen.
About Pathway Plot
Copyright ©
Anders Broe Bendtsen 1999
This program was developed
during the PhD study of Anders Broe Bendtsen.
Supervisors of the project
were Professor Kim Dam-Johansen and Associate Professor Peter Glarborg.
The PhD project was made
under the CHEC research programme at the Dept. of Chemical Engineering at the
Technical University of Denmark. (E-mail: chec@kt.dtu.dk)
Pathway Plot is provided
for public use, provided that you register with the authors, and that credit is
given by referring to the following paper:
Bendtsen, A.B.; Glarborg, P., and Dam-Johansen, K.: "Visualization methods
in analysis of detailed chemical kinetics modelling." Comp. Chem. 25, 161,
2001
Last updated: February 16th
2001
